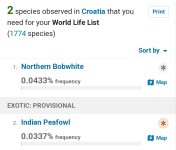
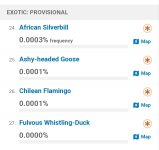
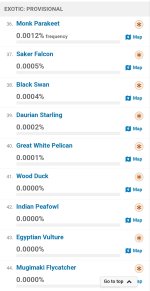
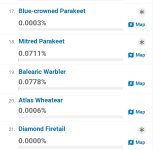
Sooo much wish they were not so opinionated wrt taxonomy. There are probably a few taxa which don't nest (=taxon definition in one taxonomy overlaps with taxon in another), but broadly, if you define things at a low enough level (subspecies, perhaps form) then you should be able to construct any of the major species-level taxonomies from those records. And that's the right way to do it.
Otherwise you get nonsense like this:
https://ebird.org/species/blkkit3?siteLanguage=en_GB
[there is a broadly correct distribution map, but the count for ebird records is zero—as is my personal count even though I recently saw loads.] This is for a well-supported split which will doubtless get ported to ebird / Clements eventually.
If conducting science, ideally I'd want to know that my analysis is robust to differences in taxonomic opinion / [future] changes in taxonomy through time. One way to do this is to repeat the analysis using different taxonomies (Clements, IOC etc) and see if I get the same results. Can't easily do with ebird...
avibase is better...